Sample name: K562_PRO-seq_40
Looper stats summary
| sample_name |
K562_PRO-seq_40 |
| sample_desc |
40% subsample of K562 PRO-seq |
| treatment |
40% subsample |
| protocol |
PRO |
| organism |
human |
| read_type |
SINGLE |
| umi_len |
0 |
| read1 |
/project/shefflab/data//guertin/fastq/K562_PRO_40pct.fastq.gz |
| srr |
K562_PRO_40pct |
| File_mb |
14593.59 |
| Read_type |
SINGLE |
| Genome |
hg38 |
| Raw_reads |
198648534 |
| Fastq_reads |
198648534 |
| Reads_with_adapter |
169233215.0 |
| Uninformative_adapter_reads |
4973679.0 |
| Pct_uninformative_adapter_reads |
2.5038 |
| Trimmed_reads |
193674855 |
| Trim_loss_rate |
2.5 |
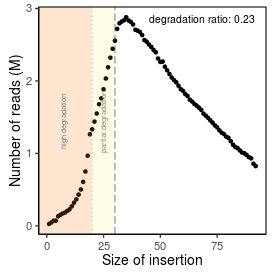
| Peak_adapter_insertion_size |
34 |
| Degradation_ratio |
0.2312 |
| Aligned_reads_human_rDNA |
17873691.0 |
| Alignment_rate_human_rDNA |
9.23 |
| Mapped_reads |
173642751 |
| QC_filtered_reads |
18693246 |
| Aligned_reads |
154949505 |
| Alignment_rate |
80.0 |
| Total_efficiency |
78.0 |
| Read_depth |
13.26 |
| Mitochondrial_reads |
3660275 |
| Maximum_read_length |
100 |
| Genome_size |
3099922541 |
| NRF |
0.53 |
| PBC1 |
0.74 |
| PBC2 |
5.58 |
| Unmapped_reads |
2158413 |
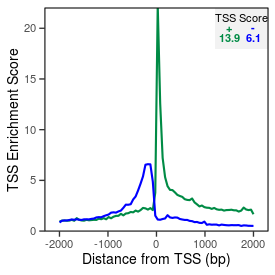
| TSS_coding_score |
13.9 |
| TSS_non-coding_score |
5.8 |
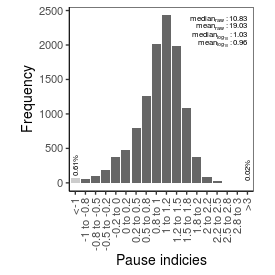
| Pause_index |
10.83 |
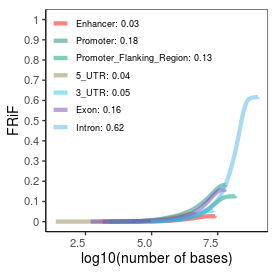
| Plus_FRiP |
0.36 |
| Minus_FRiP |
0.34 |
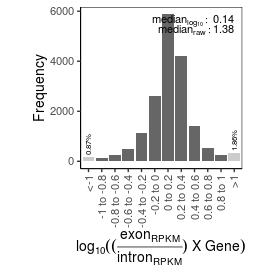
| mRNA_contamination |
1.38 |
| Time |
5:38:58 |
| Success |
06-15-12:49:59 |