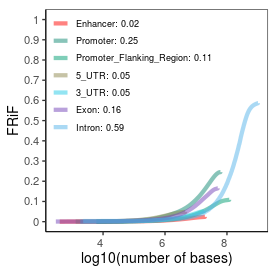
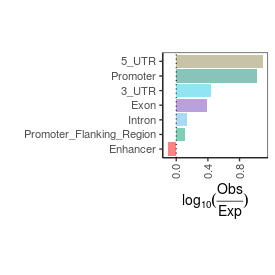
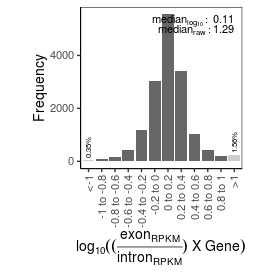
Sample name: HEK_ARF_PRO-seq
Looper stats summary
| sample_name |
HEK_ARF_PRO-seq |
| sample_desc |
HEK w/ osTIR1, ZNF143AID, ARF PRO-seq |
| treatment |
Auxin |
| protocol |
PRO |
| organism |
human |
| read_type |
PAIRED |
| umi_len |
8 |
| read1 |
/project/shefflab/data//guertin/fastq/SRR8608070_PE1.fastq.gz |
| read2 |
/project/shefflab/data//guertin/fastq/SRR8608070_PE2.fastq.gz |
| srr |
SRR8608070 |
| File_mb |
1302.45 |
| Read_type |
PAIRED |
| Genome |
hg38 |
| Raw_reads |
53049272 |
| Fastq_reads |
53049272 |
| Reads_with_adapter |
21262101.0 |
| Uninformative_adapter_reads |
8863359.0 |
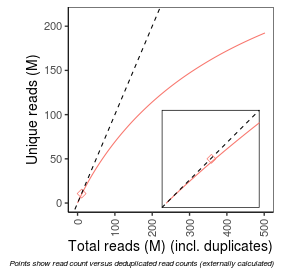
| Duplicate_reads |
778059.0 |
| Pct_uninformative_adapter_reads |
33.4156 |
| Trimmed_reads |
32625384 |
| Trim_loss_rate |
38.5 |
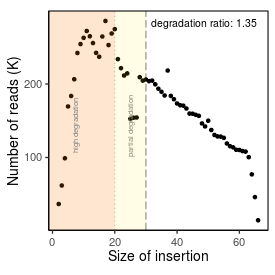
| Peak_adapter_insertion_size |
17 |
| Degradation_ratio |
1.349 |
| Aligned_reads_human_rDNA |
3622152.0 |
| Alignment_rate_human_rDNA |
11.1 |
| Mapped_reads |
24911965 |
| QC_filtered_reads |
15309473 |
| Aligned_reads |
9602492.0 |
| Alignment_rate |
29.43 |
| Total_efficiency |
18.1 |
| Read_depth |
2.05 |
| Mitochondrial_reads |
252493 |
| Maximum_read_length |
38 |
| Genome_size |
3099922541 |
| Frac_exp_unique_at_10M |
0.9164 |
| NRF |
1.0 |
| PBC1 |
5434839.0 |
| PBC2 |
5434839.0 |
| Unmapped_reads |
3997391 |
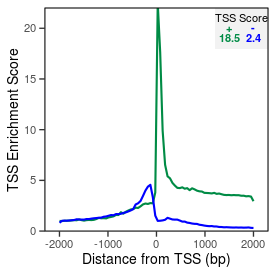
| TSS_coding_score |
18.5 |
| TSS_non-coding_score |
2.4 |
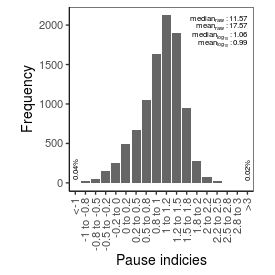
| Pause_index |
11.57 |
| Plus_FRiP |
0.42 |
| Minus_FRiP |
0.39 |
| mRNA_contamination |
1.29 |
| Time |
1:51:34 |
| Success |
06-15-09:08:45 |